This pipeline performs consensus assembly and variant calling for amplicon sequencing data (Illumina or Oxford Nanopore) generated using the ARTIC protocol. The user can specify a primer set to be used for trimming alignments; this is assumed to be ARTIC V4.1 if not specified.
The pipeline takes in a single FASTQ file (interleaved if Illumina) and processes it as follows:
- Map reads to the Wuhan-Hu-1 reference and trim ARTIC primer sequences
- Generate a consensus sequence (bcftools for Illumina; medaka for Oxford Nanopore)
- Call variants with a limit of 200x coverage, as recommended by the ARTIC network. While indels and SNVs are reported for Illumina data, only SNVs are reported for Oxford Nanopore based on benchmarking studies that indicate
small indel detection is unreliable. - Assign Pangolin and Nextclade lineages
- Predict amino acid mutations
- Predict consequences of compound variants (ex: adjacent SNVs on the same codon; frame-shifting indels followed by frame-restoring indels)
Rigorous quality checks are implemented throughout the pipeline, including flagging of variants in low complexity regions for error-prone Oxford Nanopore data and conservative lineage calls (no lineage assignments will be reported if the consensus sequence has too many N’s or is too fragmented).
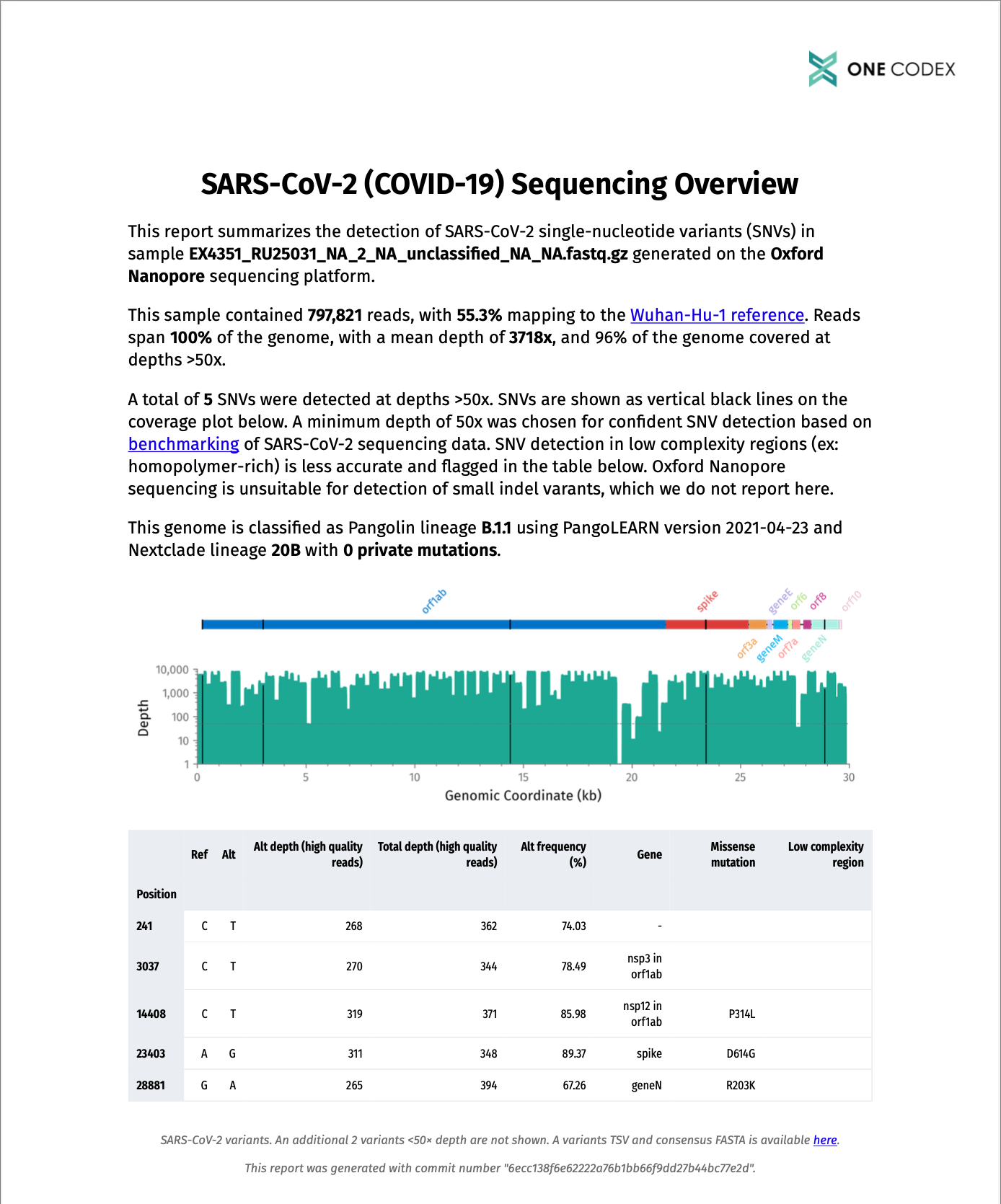
In addition to a results JSON, a PDF report is generated for each sample that tells you at a glance whether primer dropout has occurred, which amino acid mutations are present, and whether the sample contains a variant of concern. An example report is shown below.
docker build -t covid19 .Run the pipeline in the Docker image (note that fastq files are stored in git lfs so you may need to git lfs pull before executing):
docker \
run \
--rm \
--workdir /data \
--volume `pwd`:/data \
--entrypoint /bin/bash \
--env ONE_CODEX_REPORT_FILENAME=report.pdf \
--env INSTRUMENT_VENDOR=Illumina \
--env ARTIC_PRIMER_VERSION=4.1 \
covid19 \
jobscript.sh \
data/twist-target-capture/RNA_control_spike_in_10_6_100k_reads.fastq.gzFor Oxford Nanopore:
docker \
run \
--rm \
--workdir /data \
--volume `pwd`:/data \
--entrypoint /bin/bash \
--env ONE_CODEX_REPORT_FILENAME=report.pdf \
--env INSTRUMENT_VENDOR="Oxford Nanopore" \
--env ARTIC_PRIMER_VERSION=4.1 \
covid19 \
jobscript.sh \
data/twist-target-capture/RNA_control_spike_in_10_6_100k_reads.fastq.gzTo run tests, run pytest.
The requirements.txt file lists dependencies for quickly running some golden output tests across a variety of datasets. This repository is set up to use Github Actions to automatically build the Docker image and run these tests, to ensure that parameter and pipeline changes don't affect variant calls or consensus sequence generation.
Currently, integration tests are run on:
- Simulated Illumina data from the SARS-CoV-2 reference including simulated variants across the genome
- Example Twist hybrid capture data (Illumina)
- Example ARTIC v1 amplicon sequencing data (Illumina)
It also uses pre-commit to keep things clean and orderly. To get started, first install the requirements (Python 3 required): pip install -r requirements.txt. Then install the pre-commit hooks: pre-commit install --install-hooks.
Many thanks are due across the community, including but not limited to:
- @tseemann, @gkarthik, @nickloman, and many others for quick discussions on optimal SNP calling for both amplicon (ARTIC primers) and non-amplicon sequencing approaches
- @nickloman, @joshquick, @rambaut, @k-florek and others working on the ARTIC protocol for SARS-CoV-2
- @pangolin and @nextclade for surveillance tools
- Voigt lab for dnaplotlib