New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
ENH: random: Add the multivariate hypergeometric distribution. #13794
Conversation
d221568
to
85d360c
Compare
|
This is the updated version of #8056. |
85d360c
to
e9d9742
Compare
|
The refguide check doesn't like the blank line in the output of this example in the docstring: The error reported in the refguide check on the azure macOS build is It looks like the checker ignores the part of the array that occurs after the blank line. Is this is a known issue? Is there a work-around? EDIT: Worked around: I removed the blank line from the docstring. |
9e01f26
to
c838dcb
Compare
|
LGTM C/C++ passed. 🎉 🎆 😀 I don't know if that is just luck or if the LGTM folks have fixed something. Anyway, it's nice to see the green check mark. |
e350e65
to
570ea89
Compare
07ec3c3
to
f0753d7
Compare
|
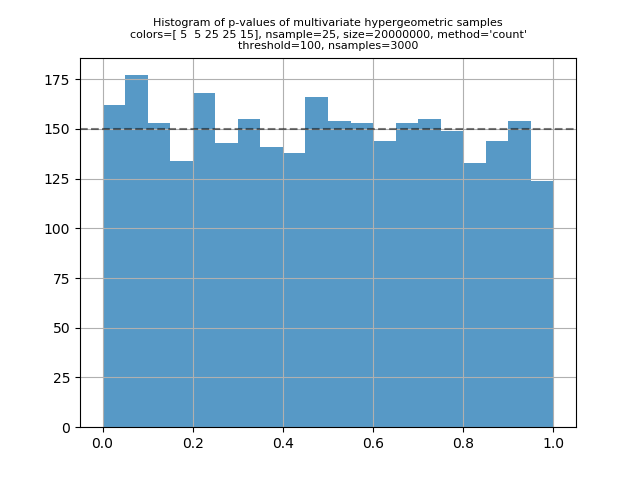
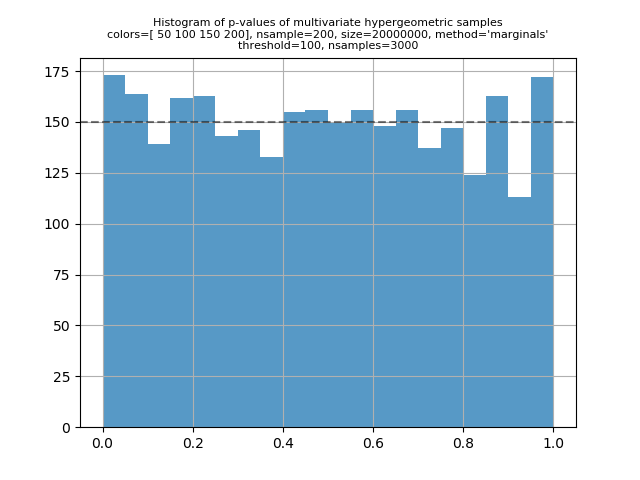
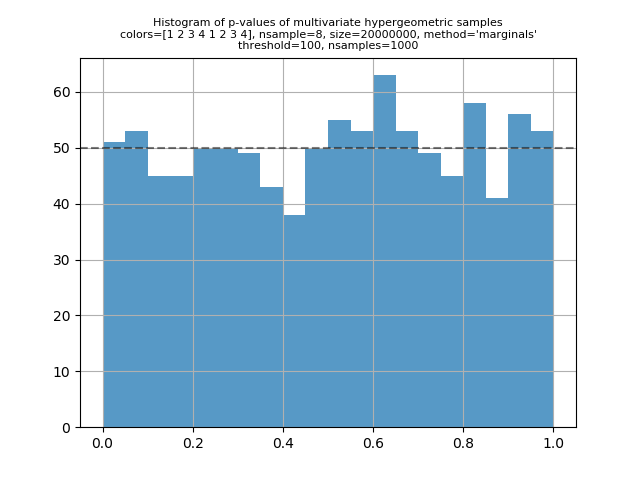
I've run some validation scripts for the multivariate hypergeometric distribution. For a given set of parameters and sample size, the basic calculation is to compute the expected counts for each point in the support of the distribution, and then do a G-test to compare that to the observed counts in a sample of that size. Under the usual statistical assumptions of the G-test, we expect the p-value to be uniformly distributed on [0, 1] if I put the scripts in my mvhg_validation repository. The two main scripts are "check1.py" and "check2.py". "check1.py" is for parameters where the both "check2.py" can handle bigger input values (but the inputs still can't be too big if you want the calculation to complete in a reasonable amount of time). The script will aggregate points in the support where the expected value is less than 100 by grouping points such that the sum of the expected value in each group is at least 100. I've been running "check2.py" so that it generates one to three thousand p-values. The script plots a histogram of the p-values. A visual inspection can confirm that the distribution of the p-values looks uniform. Here's a typical output printed by "check1.py": And here are several plots generated by "check2.py": |
f0753d7
to
b0b9c33
Compare
|
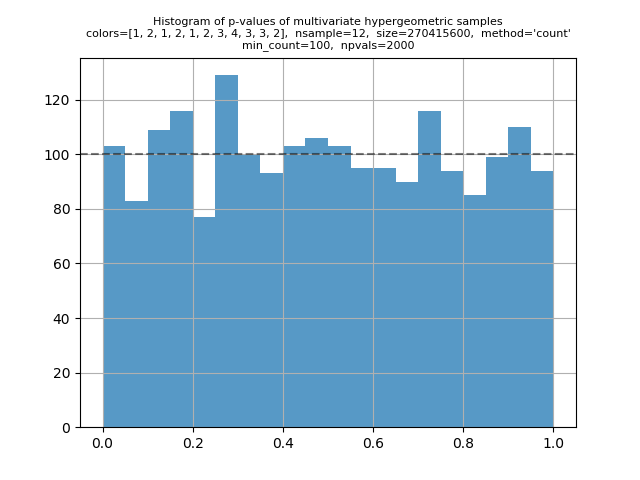
One more p-value histogram, using |
|
This is ready for review. (Reminder: this is the updated version of #8056.) |
|
A friendly request: could someone remove the "WIP" tag from this pull request? It is ready for review. I know that the folks who would likely review this are focused on finishing the randomgen update, so I don't expect a review to happen soon, but I want to be sure that someone who does find time doesn't pass over this one because of the "WIP" tag. |
|
Thanks @seberg! |
|
@WarrenWeckesser there is a merge conflict. |
|
The pull request includes two methods for generating a sample:
It is not in the pull request, but I also experimented with a third implementation that used Floyd's algorithm:
It avoided creating the (potentially big) array that is created by the "count" method. I used the C library khash for the hash table. This third method is not in the pull request because, in all my tests, either the "count" method or the "marginals" method was faster. After seeing the pull request "MAINT: Rewrite Floyd algorithm" (#13812), I'm inspired to experiment a bit more with the hash table method. I only timed the full process of generating samples, so I don't know if swapping out khash for the simple hash table with linear probing would make a signifcant difference. |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The code generally LGTM. master needs to be merged in and the tests adapted to the current style.
Whether this goes into 1.17, I leave up to @charris.
|
@WarrenWeckesser ping |
|
It is a shame this nice addition didn't make it into 1.17. @WarrenWeckesser will you be picking it up again? |
|
Yes, I will get back to this in a few weeks, if not sooner. |
44ca481
to
e29f9c1
Compare
|
Rebased and updated. |
e29f9c1
to
244270d
Compare
|
@rkern's last comment was
I have since rebased and updated the tests. For anyone taking a fresh look at this PR, the essential additions are:
All the other changes are to support the above, add tests, or update the documentation. |
|
xref #14608, since both that and this should be merged before 1.18, and both touch generator.pyx |
The new method
multivariate_hypergeometric(self, object colors, object nsample,
size=None, method='marginals')
of the class numpy.random.Generator implements the multivariate
hypergeometric distribution; see
https://en.wikipedia.org/wiki/Hypergeometric_distribution,
specifically the section "Multivariate hypergeometric distribution".
Two algorithms are implemented. The user selects which algorithm
to use with the `method` parameter. The default, `method='marginals'`,
is based on repeated calls of the univariate hypergeometric
distribution function. The other algorithm, selected with
`method='count'`, is a brute-force method that allocates an
internal array of length ``sum(colors)``. It should only be used
when that value is small, but it can be much faster than the
"marginals" algorithm in that case.
The C implementations of the two methods are in the files
random_mvhg_count.c and random_mvhg_marginals.c in
numpy/random/src/distributions.
244270d
to
8634b18
Compare
|
I updated the PR to match the changes in #14608. I don't know why the Azure tests didn't run. |
|
no idea why azure is not running :( |
|
Thanks @WarrenWeckesser |

The new method
of the class numpy.random.Generator implements the multivariate
hypergeometric distribution; see
https://en.wikipedia.org/wiki/Hypergeometric_distribution,
specifically the section "Multivariate hypergeometric distribution".
Two algorithms are implemented. The user selects which algorithm
to use with the
methodparameter. The default,method='marginals',is based on repeated calls of the univariate hypergeometric
distribution function. The other algorithm, selected with
method='count', is a brute-force method that allocates aninternal array of length
sum(colors). It should only be usedwhen that value is small, but it can be much faster than the
"marginals" algorithm in that case.
The C implementations of the two methods are in the files
random_mvhg_count.c and random_mvhg_marginals.c in
numpy/random/src/distributions.
The new file numpy/random/src/distributions/util.c contains the
function safe_sum_nonneg_int64() for summing nonnegative int64
values while checking for overflow.