-
Notifications
You must be signed in to change notification settings - Fork 128
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
3dmol format for visualization in jupyter (#357)
Example: https://nbviewer.org/gist/njzjz/14a638d1b5c85c1b2a4882418c34dd9d 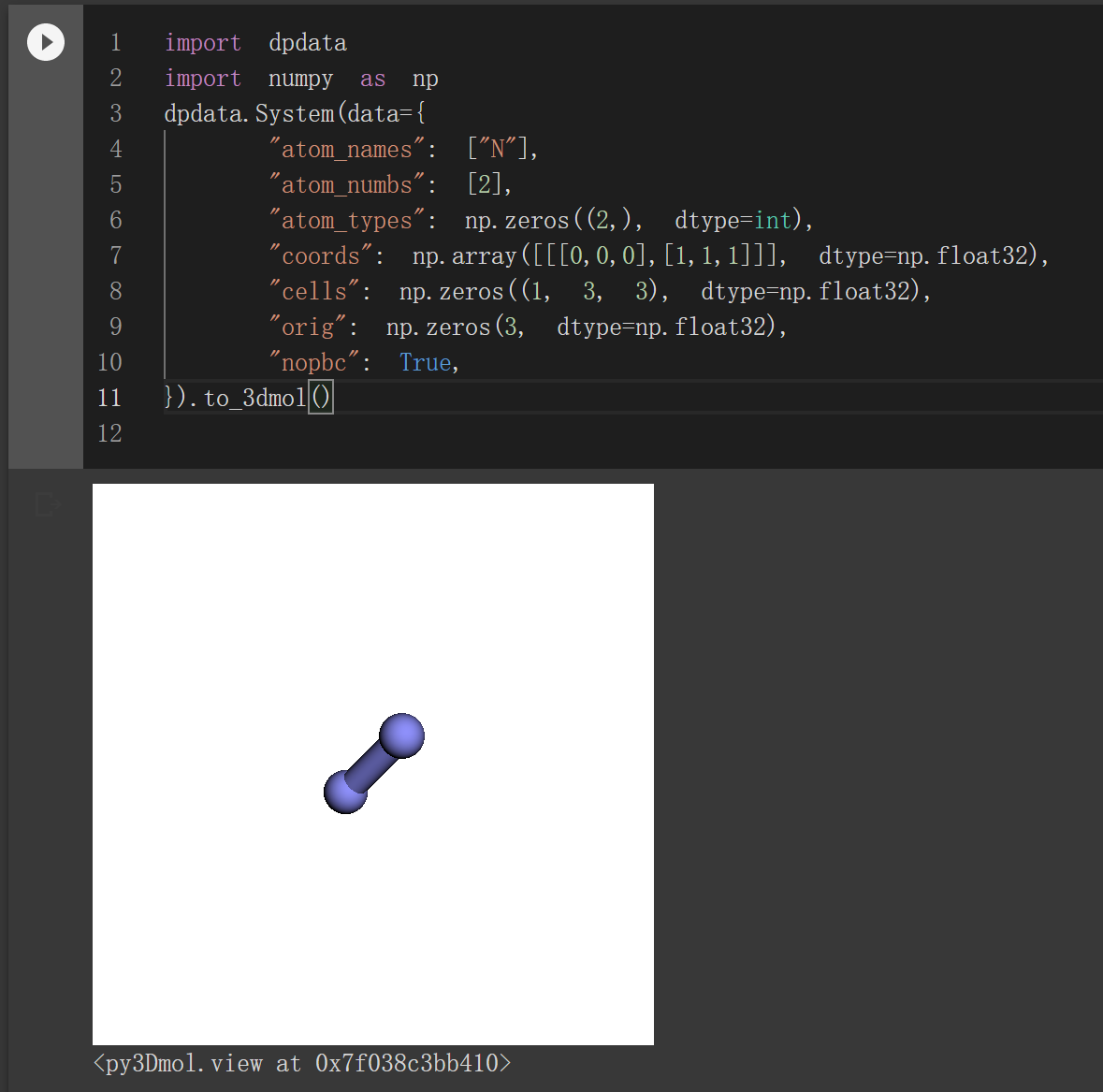
- Loading branch information
Showing
1 changed file
with
44 additions
and
0 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,44 @@ | ||
| from typing import Tuple | ||
| import numpy as np | ||
|
|
||
| from dpdata.format import Format | ||
| from dpdata.xyz.xyz import coord_to_xyz | ||
|
|
||
|
|
||
| @Format.register("3dmol") | ||
| class Py3DMolFormat(Format): | ||
| """3DMol format. | ||
| To use this format, py3Dmol should be installed in advance. | ||
| """ | ||
| def to_system(self, | ||
| data: dict, | ||
| f_idx: int = 0, | ||
| size: Tuple[int] = (300,300), | ||
| style: dict = {"stick":{}, "sphere":{"radius":0.4}}, | ||
| **kwargs): | ||
| """Show 3D structure of a frame in jupyter. | ||
| Parameters | ||
| ---------- | ||
| data : dict | ||
| system data | ||
| f_idx : int | ||
| frame index to show | ||
| size : tuple[int] | ||
| (width, height) of the widget | ||
| style : dict | ||
| style of 3DMol. Read 3DMol documentation for details. | ||
| Examples | ||
| -------- | ||
| >>> system.to_3dmol() | ||
| """ | ||
| import py3Dmol | ||
| types = np.array(data['atom_names'])[data['atom_types']] | ||
| xyz = coord_to_xyz(data['coords'][f_idx], types) | ||
| viewer = py3Dmol.view(width=size[0], height=size[1]) | ||
| viewer.addModel(xyz, 'xyz') | ||
| viewer.setStyle(style.copy()) | ||
| viewer.zoomTo() | ||
| return viewer |