Implemented a similar approach described in the paper by Kermany et al., “Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning.” [1]
The network consists of a ResNet152 [6] with pretrained weights and all but the last 3 layers frozen, as well as an adjusted fully connected layer for the number of classes. Initial model trained with an NVIDIA 1080ti GPU, 16GB RAM, and a Ryzen 1800X.
Experimented with smaller, simpler convolutional networks based on the work of [3], but ResNet152 transfer learning was more successful.
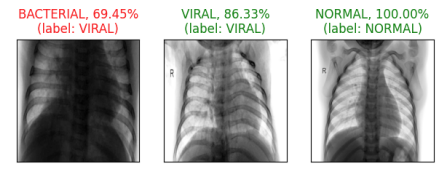
- Achieved 85.1% overall accuracy, 80.7% weighted f1-score after 30 epochs, a batch size of 32, and a learning rate of 1e-4.
To prepare the data, locate the data from the Mendeley Medical Chest X-Ray dataset [2] and create a test/train split, subdivided into normal or pneumonia cases. I used the data split from reference [4]. A PyTorch backend is required.
- Run main.py with the data path specified organized into test/train folders, with the example directory structure given below. A data path is required. Example:
python3 main.py --data_path=./data
- To monitor training, run
tensorboard --logdir=tflogs_dir
-
Training initial iteration of model
- Data augmentation, testing, hyperparameter optimization
-
Packaging and Serving the Model
- Create torch model archive for serving
DeepTransfer
├── data
│ ├── custom_dataset.py
│ ├── __init__.py
│ └── raw
│ ├── test
│ │ ├── NORMAL
│ │ │ ├── IM-0574-0001.jpeg
│ │ │ └── NORMAL2-IM-1049-0001.jpeg
│ │ └── PNEUMONIA
│ │ └── person1372_bacteria_3499.jpeg
│ └── train
│ ├── NORMAL
│ │ ├── IM-0041-0001.jpeg
│ │ └── NORMAL2-IM-0329-0001.jpeg
│ └── PNEUMONIA
│ ├── person154_bacteria_728.jpeg
│ └── person16_virus_47.jpeg
├── docs
│ └── tensorboard_util.png
├── logs
├── main.py
├── model
│ └── checkpoints
├── model_store
├── README.md
├── requirements.txt
└── utils
├── custom_dataset.py
├── data_utils.py
└── __init__.py
- Kermany, D. S., Goldbaum, M., Cai, W., Valentim, C. C. S., Liang, H., Baxter, S. L., McKeown, A., Yang, G., Wu, X., Yan, F., Dong, J., Prasadha, M. K., Pei, J., Ting, M. Y. L., Zhu, J., Li, C., Hewett, S., Dong, J., Ziyar, I., … Zhang, K. (2018). Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell, 172(5), 1122–1131.e9. https://doi.org/10.1016/j.cell.2018.02.010
- Kermany, D., Zhang, K., & Goldbaum, M. (2018). Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images for Classification. 2. https://doi.org/10.17632/rscbjbr9sj.2
- Understanding Transfer Learning for Medical Imaging. (2019). Google AI Blog. Retrieved May 15, 2020, from http://ai.googleblog.com/2019/12/understanding-transfer-learning-for.html
- Chest X-Ray Images (Pneumonia). (2019). https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia
- Visualizing Models, Data, and Training with TensorBoard. (2017). https://pytorch.org/tutorials/intermediate/tensorboard_tutorial.html
- He, K., Zhang, X., Ren, S., & Sun, J. (2015). Deep Residual Learning for Image Recognition. arXiv:1512.03385 [Cs]. http://arxiv.org/abs/1512.03385